Evolution
and the Immune System
Sean
D. Pitman M.D.
©September
2006
 Everyone
has experienced sickness at one time or another in his or her life.
However, most people get better and often do not get that same "bug" again.
Why is this? There are
literally millions of "bugs" and other things in our
everyday environment that can make us sick.
Why then do we generally remain so healthy?
The reason is because most of us have a highly effective immune system.
A healthy immune system is very good at searching out and killing foreign
invaders that can make the body sick. However, if the immune system is not
functioning well, there will certainly be a lot of problems.
Everyone
has experienced sickness at one time or another in his or her life.
However, most people get better and often do not get that same "bug" again.
Why is this? There are
literally millions of "bugs" and other things in our
everyday environment that can make us sick.
Why then do we generally remain so healthy?
The reason is because most of us have a highly effective immune system.
A healthy immune system is very good at searching out and killing foreign
invaders that can make the body sick. However, if the immune system is not
functioning well, there will certainly be a lot of problems.
As
an example of this, consider the famous HIV virus (Human Immunodeficiency
Virus). This virus causes the disease known as AIDS (Acquired
Immune Deficiency Syndrome). A
person with AIDS has a very weakened immune system.
Because of a lack of immunity, infections plague the AIDS victim that do
not normally infect humans. Interestingly enough, it is not the HIV virus
itself that makes these ![]() people sick. The HIV virus specifically attacks
immune cells called T-helper cells (click on the video illustration - i.e., the
black box). When these cells are lost in high
enough numbers, the immune system starts to shut down. The symptoms of
AIDS are the result of a loss of immunity. With the loss of immunity, a
host of infectious processes start to overwhelm the body. Eventually,
these infections kill their unfortunate host.
people sick. The HIV virus specifically attacks
immune cells called T-helper cells (click on the video illustration - i.e., the
black box). When these cells are lost in high
enough numbers, the immune system starts to shut down. The symptoms of
AIDS are the result of a loss of immunity. With the loss of immunity, a
host of infectious processes start to overwhelm the body. Eventually,
these infections kill their unfortunate host.
Now
that we understand just how important it is to recognize bad "bugs",
how do our bodies become capable of recognizing the millions and billions and
even trillions of different arrangements of things that can make us sick?
Well, it is through a
Darwinian-style selection process where the fittest survive and the
weakest die. In our bodies we have
cells that are specialized immune cells called T-cells. They
go to "school" to learn the difference between "self" and
"non-self". Certainly one would not want his/her own immune system to
attack his/her own body! Sometimes this does happen and it is referred to as
an autoimmune disease. However, normally, the T-cells are educated in
a very tough school so that they do not attack one's own body or
"self". But
how exactly are they trained to recognize the difference between self and
non-self?
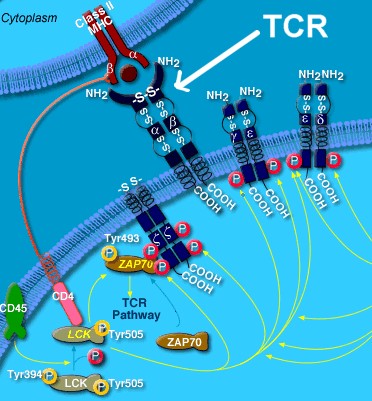 Well,
T-cells are capable of being able to tell the difference between certain molecules (antigens) that are
presented (by MHC class I molecules) on
the surfaces of all "self" cells in the body that they are supposed to
protect compared to all foreign or "abnormal" antigens that are
associated with outside invaders or internal disease processes (like cancer). For
example, when an outside invader, like a bacterium, enters the body, other cells
(macrophages) may catch this invader and "eat" it. The
macrophage then presents small parts of the invader on its own surface in
association with a certain molecule called MHC class II. If a cell within
the body gets "sick" or diseased (i.e., cancerous), certain molecules
that are usually hidden from the immune system may be produced. These are
now presented on the surface of this sick cell in association with MHC class I
molecules. T-cells recognize these new molecules as "non-self"
and kill the sick or diseased cell. Once the T-lymphocyte recognizes an
infected cell, it produces a set of new proteins that it places on the surface
of the sick or diseased cell. Those proteins then bind to receptors on the
infected cell called "death domain receptors" (including the Fas
ligand and Trail receptors). This binding triggers a cascade of events in the
infected cell that leads to cell suicide, called apoptosis.
Well,
T-cells are capable of being able to tell the difference between certain molecules (antigens) that are
presented (by MHC class I molecules) on
the surfaces of all "self" cells in the body that they are supposed to
protect compared to all foreign or "abnormal" antigens that are
associated with outside invaders or internal disease processes (like cancer). For
example, when an outside invader, like a bacterium, enters the body, other cells
(macrophages) may catch this invader and "eat" it. The
macrophage then presents small parts of the invader on its own surface in
association with a certain molecule called MHC class II. If a cell within
the body gets "sick" or diseased (i.e., cancerous), certain molecules
that are usually hidden from the immune system may be produced. These are
now presented on the surface of this sick cell in association with MHC class I
molecules. T-cells recognize these new molecules as "non-self"
and kill the sick or diseased cell. Once the T-lymphocyte recognizes an
infected cell, it produces a set of new proteins that it places on the surface
of the sick or diseased cell. Those proteins then bind to receptors on the
infected cell called "death domain receptors" (including the Fas
ligand and Trail receptors). This binding triggers a cascade of events in the
infected cell that leads to cell suicide, called apoptosis.
T-cells sense or
"feel" these antigen molecules with their own fairly tall Y-shaped molecules called T-cell
receptors (like little hands on the ends of long arms). These
T-cell receptors (TCRs) are analogous to wider Y-shaped antibody molecules (also known as "immunoglobulins") produce by
B-cells (see illustrations below). However, TCRs exist only in
a membrane-bound form. They are able to carry out their particular
function without the need to leave the cell surface. Their receptors are
used for the detection of foreign antigens, and do not directly mediate an
effector response (except in the case of specialized "cytotoxic"
T-cells).4
Those
T-cells that do not recognize self-antigens as being part of the body are
killed off before they get out of school (and this is a rather large majority of
the "students" that enter this school).
It is a tough school indeed if the students do not learn their lessons!
Those cells that do recognize self-antigens as "self" graduate to go and search the
body for non-self antigens to attack and eliminate.
Each
educated T-cell has only one type of receptor so only one specific antigen
can be recognized. But, how many possible
antigens are there? "The total number of possible epitopes is,
therefore, 20B since there are 20 different amino acids."5
Well, the typical length of an antigen epitope ("B" in the preceding
formula) is about 20 amino acid residues.5 So, the total number
of possible antigen epitopes is about 2020 or 104,857,600,000,000,000,000,000,000
or ~100 trillion trillion.
Since there are
trillions of different possible antigen epitopes, how does one's immune
system cope with such a variety of potential enemies?
Well, there are many immune cells produced by the body. In humans,
in particular, about 1012 lymphocytes are present at any given time.6
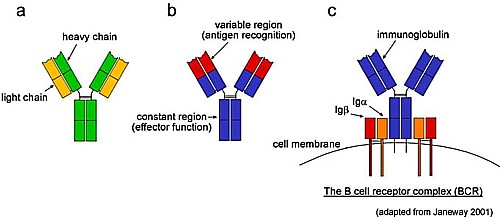 Not all the T-cells have different Y-shaped receptors,
but many of them do. Chances
are that if enough non-self enemies get into the body at least one of the immune
cells will recognize the non-self marker sequences or "antigens" located on this invader as
"foreign" to at least some useful degree.
The odds that a single T-cell will recognize a random epitope to at least some
useful degree is about 1 in 1012. So, does this mean it would
take a trillion different T-cells to cover all possible invaders? Well,
no. The reason is because an average cell or foreign invader
"bug" has about 1012 different epitopes. So, on
average, a single T-cell will recognize at least one of the potential antigen
epitopes of a foreign invader.5
Not all the T-cells have different Y-shaped receptors,
but many of them do. Chances
are that if enough non-self enemies get into the body at least one of the immune
cells will recognize the non-self marker sequences or "antigens" located on this invader as
"foreign" to at least some useful degree.
The odds that a single T-cell will recognize a random epitope to at least some
useful degree is about 1 in 1012. So, does this mean it would
take a trillion different T-cells to cover all possible invaders? Well,
no. The reason is because an average cell or foreign invader
"bug" has about 1012 different epitopes. So, on
average, a single T-cell will recognize at least one of the potential antigen
epitopes of a foreign invader.5
When this happens this particular
T-cell sounds the alarm that the
body has been invaded. Other immune cells,
called B-cells, are also activated, but only those that specifically produce
antibodies that have a pretty good match to the foreign antigen epitope expressed by the
invading organism. The invader,
with its non-self antigens, is attacked. However,
if only a few immune cells recognize the invader upon initial exposure, the
initial attack might be rather weak. The
resulting sickness may
linger on for some time before the body can kill off the offending
invader. The good thing is that the
immune system remembers this particular invader for the future so it can kill
the invader more quickly if it ever sees its particular antigen marker again.
But how does this memory work?
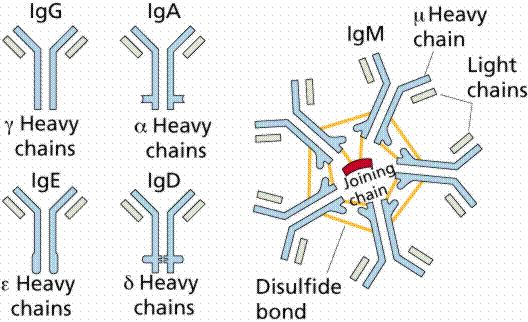 The
B-cell that recognized the foreign antigen clones itself to make many
nearly identical copies of itself - with slight variations.
Now, there are many B-cells that will recognize this particular
foreign antigen. If infected again
by an invader with this particular antigen, the immune system is ready and
produces many more specific antibodies than before. This kills
the invader much more quickly - making the body "immune" to this particular bug.
The
B-cell that recognized the foreign antigen clones itself to make many
nearly identical copies of itself - with slight variations.
Now, there are many B-cells that will recognize this particular
foreign antigen. If infected again
by an invader with this particular antigen, the immune system is ready and
produces many more specific antibodies than before. This kills
the invader much more quickly - making the body "immune" to this particular bug.
This
is how vaccines work. A vaccine
presents the body with the antigens of either a dead or a weakened bug.
In this way the body can prepare to kill that particular bug without
first having to go through the sickness that the bug may causes.
Programmed
Variability
Many
people, including scientists, claim that this process is evolution in
action.3,4,5 Is this actually true?
After all, this system does use survival of the fittest and a
function-based selection process to create an incredible diversity of immune
cells. Is this not evolution in
action? In
thinking about this question, remember that the immune system had no
initial knowledge about all the evil antigens in the world or just which ones it
might have to combat. So, how did
it get its gigantic arsenal of options?
Well,
the initial production of variety of T-cell receptors and B-cell produced
antibodies is done by purely random mutation without any function-based
selection. However, subsequent refinements of immune system function are
indeed based on both random mutations and function-based selection over several
generations of B-cells.
This may be a form of evolution in action,
but it isn't quite what many evolutionists think it is. Lets
look into this process in a bit more detail.
Antibody
Structure
Antibodies are proteins and so they are coded for by DNA.
Antibodies are Y-shaped molecules with two different protein strands
called heavy and light chains. At
the tips of the V-ends of the Y there are "variable regions" on both the
light and heavy chains that can be different from cell to cell.
Also, within this variable region are half a dozen or so "hypervariable
regions". The rest of the antibody does not vary in its
protein sequencing from other antibodies within the body.
Each one of the two chains (heavy and light) are coded for by specific
sections of DNA. Each section of
DNA can code for part of the final antibody for that cell if it becomes an
immune cell. Even though there are
many options for each section only one option will be chosen for a particular
section. This choice is determined
by the random recombination of one option for one section with one option for
each one of the other sections (see illustration).
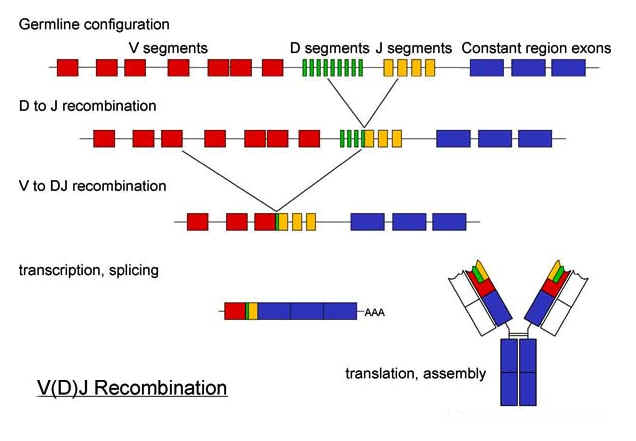 For
light chains there are about 250 V-segments (or separate options in the original
DNA), four J-segments, and three different ways that a V-segment can join to a
J-segment when the DNA is spliced. The
final product for a light chain in DNA is one V-segment followed by a single
J-segment. This makes up the "variable region" of the light chain. The
constant region of DNA for the light chain follows this region.
The rest of the gene options are discarded for that cell.
For
light chains there are about 250 V-segments (or separate options in the original
DNA), four J-segments, and three different ways that a V-segment can join to a
J-segment when the DNA is spliced. The
final product for a light chain in DNA is one V-segment followed by a single
J-segment. This makes up the "variable region" of the light chain. The
constant region of DNA for the light chain follows this region.
The rest of the gene options are discarded for that cell.
For
the heavy chains, there are about 250 V-segments, 15 D-segments, and 5
J-segments - followed by the constant region.
Just like in the light chain DNA, only one option from each segment type
is chosen for the final splicing of DNA so that just one V-option is followed by
one D-option which is followed by one J-option.
This makes up the "variable region" of the heavy chain.
The constant region follows just like in the light chain.
The
large number of different options and their different possible combinations give
rise to the huge variety of antibody possibilities.
This number can be roughly calculated as follows:
Light chains have 250 V-segments, 4 J-segments, and 3 possible joining
frames. This gives a total of 250 x
4 x 3 or 3000 different kinds of complete light chain possibilities.
Heavy chains have 250 V-segments, 15 D-segments, 5 J-segments, and 3
different joining frames. This
gives a total of 250 x 15 x 5 x 3 or 56,250 different kinds of complete heavy
chain possibilities. Combining the
chains gives around 1.7 x 108 (~170 million) different possible
antibody arrangements. Adding in the various hypervariable differences
within the variable regions gives around 1 x 1010 (~10 billion)
different antibody specificities.1,2
Discussion
Certainly, the potential for immune cell variety is great, but is the process
that produces this variety really an example of the Darwinian-style evolution at work?
3 According to the theory of evolution all living things evolved from a
single common ancestor over time. The
differences between the resulting life forms arose when random mutations
happened upon some sort of new beneficial function that was beneficial to a
particular life form in a particular environment or situation. More and more of these differences
added up over time until it resulted in the vast diversity of living things that
we see today. That's the theory of evolution in a nut shell. That's how it
is supposed to have worked.
Now, in thinking about the immune system, is there a parallel with the theory of
evolution? What is evolving in the immune system? The overall
structure and function of the immune cells
does not evolve over time and neither do the types of antibodies
that are produced. All the major types of immune cells and antibodies are produced
before any functional selection takes place. Only after the T-cells
are formed do they go to school to be "selected". Also, this
selection process only selects for a very limited ability - the ability to
recognize and refrain from attacking self antigens. This particular
selection process is very specific and it does not change over time to produce
anything new. So, the major differences in antibody specificity that are
maintained were all made before any selection process
took place. As far as the immune system is concerned, the initial action
of "selection" and survival of the fittest only narrows the
field. It reduces the antibody diversity that was initially created before
selection came along.
This is different from Darwinian-style evolution where
natural selection is supposed to be able to create diversity. Selection,
in the case of the immune system, does
not expand or create more diversity - at least not in the initial steps of
immune system education. Perhaps, then, later steps in the function of the
immune system show Darwinian-style evolution of truly novel functions?
Real
Evolution
As a review, remember that only a small fraction of the T-cells survive the
first selection step. When the body is exposed to a foreign invader with a
specific antigenic marker sequence, T-cells that match that marker stimulate
B-cells that also match to reproduce themselves - in a clonal fashion. If just any immune cell was allowed to clone or
reproduce itself, the immune system would not be nearly as
effective. However, mass reproduction of nearly identical copies of a B-cell that did in fact recognize a specific type of foreign invader, is
very helpful in preventing future sickness by that particular invader should it ever invade the body again.
Interestingly though, once a particular B-cell is selected for mass
replication, the offspring of that parent cell are not exactly the same.
The antibodies that are produced by the offspring B-cells are slightly
different - usually in their "hypervariable regions".
These changes are indeed random changes that were not present in
the original pool of immune system options. In other words, they are truly new
sequences. When the same foreign antigen is
encountered again, those slightly different clones of the original B-cell
that recognize the antigen better will be preferentially selected, over
the siblings that do not have as close a match, to be cloned and produce the
offspring of the next generation.
In this manner the specificity of immunity will indeed evolve in a stepwise
truly Darwinian-style fashion of improvement over time.
"The B-cells expressing low
affinity antibody on their surface become progressively less able to bind and be
stimulated by antigen; in the environment of the germinal center, these poorly
stimulated B cells are programmed to die by a specific process known as
"apoptosis" (Choe et al, J Immunol 157:1006,1996). In contrast, the
cells with high affinity antibody continue to bind antigen, and thus continue be
stimulated to proliferate and secrete antibody. As the antigen concentration
progressively falls while mutation and selection continue, the intensity of the
selective pressure for high affinity increases. Repeated cycles of mutation and
selection can lead to affinity levels 100-fold higher than that of the original
unmutated antibody. The 'competition' for efficient antigen binding has been
shown to be the selective force driving the rise in antibody affinity, since if
antigen is repeatedly administered to prevent the drop in antigen level and
thereby eliminate the selective pressure for efficient antigen binding, antibody
affinity does not rise (Eisen and Siskind, Biochemistry 3:996, 1964).
Furthermore, when selection pressure has been experimentally removed by
engineering mice with impaired capacity for programmed death by apoptosis, many
B cells are found that make mutated antibodies with low affinity (Takahashi et
al. J Exp Med. 190:39, 1999).
Late in the course of an immune response, as antigen becomes
completely cleared from the bloodstream the amount of antibody secreted
gradually falls and the immune response ends; but a subset of the last group of
highly efficient cells persists as a quiescent population known as 'memory
cells,' ready to respond with rapid secretion of high affinity antibody should
they ever be triggered by another encounter with the same antigen in the
future." 3
So, basically what we have here is random mutation combined
with function-based selection that makes improvements over time. By definition,
this sounds an awful lot like real evolution in action, but there is just one
little catch.
One
Little Catch
Imagine a sequence space of all possible antibody specificities that an immune system can
make. Such a space would contain literally billions upon billions, and probably
even trillions of potential antibody specificities - - right? Now, from the perspective of a given antigen (actually an
epitope or a small portion of an antigen), there would be a continuum of reactivity if it were
brought in contact with each one of the potential antibodies in sequence space.
The continuum would range from an extremely poor fit to an extremely good fit.
Imagining this continuum as a line where, on every point of that line, there may be
a number of antibodies that would match our particular antigen to exactly the
same degree of selectability (i.e., they would activate the immune response to
the same selectable degree). Although there may be a few equivalent antibody
options at every point on our continuum, the odds are that the majority of
potential antibodies are fairly evenly spread out over the entire
continuum.
Given such a continuum as a true picture of reality, there are basically no significant neutral gaps
(with regard to the function of immunity) to cross in the evolution of antibody
specificity of a given antigen. In other words, the odds are therefore very high that a give
change/recombination/mutation to an antibody sequence will actually be
functionally different (either more or less binding affinity for a specific
antigen epitope) compared to what came before. Since a high ratio of potential
changes would be functionally different, evolution, even of highly specific
sequences, will occur rapidly. This explains the relatively rapid refinement of
antibody specificity when immune cells are repeatedly exposed to the same
antigen.
Another way to look at it is to think of the foreign antigen as a pre-formed pattern,
model, goal sequence, or type of template. The goal of creating random antibody
sequences is to change sequences that already match the antigen template to see
if random changes can keep what already matches and yet come up with additional
matches to create an even better fit. This system of evolution works
because the antigen is limited to a certain number of potential characters at
each position of its sequence, and the random antibody sequence generator is
programmed to cover all of these possibilities at each possible sequence
position.
Methinks
it is Like a Weasel
For example, say that the antigen
target or "goal" reads like Dawkins's famous, "Methinks
it is like a weasel" illustration. Starting with a bunch of random
phrases (antibodies) and mutating them at random sequence positions in each
"generation" of phrases, all that has to be done is to find out which
one matches the goal phrase, "Methinks it is like a weasel" at the
most positions and then use that "best" phrase to populate the next
generation. Obviously, if the goal is already in place ahead of time, it is easy
to evolve other phrases to match the goal phrase in short order using random
changes and sequence-based comparison - as Dawkins's illustration clearly
establishes.
Exactly the same thing happens with antibody evolution. It is already
pre-established that certain molecules will be attracted to each other. If more
of these molecules are lined up in the right places, there will be a stronger
attraction. If the immune system is programmed to recognize stronger binding as
"beneficial", stronger and stronger binding will quickly evolve with
the random changes of the sequences of each subsequent generation.
The
Non-Beneficial Gap Problem
Of course, the problem for evolutionists is clearly illustrated by Behe and many
others in their devastating critique of Dawkins's "Methinks it is like a
weasel" scenario. If all potentially beneficial types of functions could be
built by matching sequences to other pre-established sequences, then
Darwinian-style evolution of all types of functional systems would be a piece of cake. Of course, for most
functions found in operation within living things, evolution is not thought to
have worked like this at all. There simply is no pre-established sequence or
"ideal" with which to compare the evolving sequence. For many types of
complex functions, like bacterial
motility for example, there simply is no pathway of step-by-step improvements
for each and every single amino
acid change/mutation. The forces of Darwinian-style evolution run into the Non-Beneficial
Gap Problem when it comes to such higher-level functions.
Consider the function of bacterial
motility more closely for comparison. Bacterial motility will not be realized at all until a minimum
number and specified arrangement of amino acids are entirely in place.
Before such assembly is realized, there is no motility and there is no
pre-established model sequence available to guide the random changes via
function-based selection to produce such an integrated system one single step or
single character change at a time. Without such a
model sequence acting as a function-based guide directing each change toward
continual improvement, the system of function-based selection gets blinded
pretty quickly. The exponentially
expanding gaps of neutral and detrimental sequences quickly separate the
fewer and fewer potentially beneficial sequences at higher and higher levels of
functional/meaningful complexity.
It is the resulting random walk
created by the huge ocean of functionally neutral and detrimental sequences in
sequence space at higher levels of functional complexity that destroys the
evolutionary mechanism of random mutation and natural selection. The random walk
is made possible by the blindness of nature to functionally neutral sequence
changes/mutations as well as nature's aversion to functionally detrimental
changes. Because of this blindness to neutral differences and aversion to
detrimental differences in different sequences, a seemingly small
neutral/detrimental gap between potentially beneficial functions translates into
a correspondingly enormous random walk, which grows exponentially in length (by
a factor of 2) with each doubling of the average neutral gap. So, beyond the
lowest levels of functional complexity evolution simply stalls out this side of
a practical eternity of time (i.e., trillions upon trillions upon "zillions" of years of
time).
Low
Level Evolution
So, we see that even though the immune system does undergo random mutation with
function-based selection and survival of the fittest with improved fitness over
time, this is not a very close parallel to Darwinian-style evolution when it
comes to explaining the evolution of many systems of function that exist within
all living things. Most of the great
varieties within "kinds" in the animal and plant kingdoms can easily be
explained by "preprogrammed" or inherent genetic abilities for variety (such
as the phenomenon of genetic recombination as first described by Gregor
Mendel). There are certain limited
situations were new functions can and do evolve without pre-established
templates, such as in the cases of lactase
and nylonase evolution as well as antibiotic
resistance in bacteria, but these examples of evolution in action have clear
boundaries that have never been crossed. However, even with their
limitations, the evolution of antibiotic
resistance, lactase
function, computer
codes, and the like, are far better examples of evolution creating novel
sequences than is the process of directed immune system variability which does
nothing more "evolutionary" than evolve a string of characters to
match another pre-established string of characters. Such template-matching
evolution just doesn't solve the problem for the larger Theory of Evolution.
-
Stryer,
Lubert. Biochemistry,
3rd ed., 1988.
-
Janeway & Travis, Immunobiology, Garland Publishers, 4th ed. 2000.
-
Max,
Edward E., The Evolution of Improved Fitness by Random Mutation Plus
Selection, The Talk.Origins Archive, © 1999-2001 ( Link
)
-
Inlay,
Matt, Evolving Immunity - A Response to Chapter 6 of Darwin's Black
Box, The Talk.Origins Archive, July 17, 2002 ( Link
)
-
Jun
Sun, David J. Earl, and Michael W. Deem, Glassy Dynamics in the
Adaptive Immune Response Prevents Autoimmune Disease, Physical Review
Letters, 95, 148104, September 30, 2005 (Link
)
-
Niels
K. Jerne, The Generative Grammar of the Immune System, Nobel
Lecture, 8 December 1984 ( Link
)
.
Home Page
. Truth,
the Scientific Method, and Evolution
.
Methinks
it is Like a Weasel
. The
Cat and the Hat - The Evolution of Code
.
Maquiziliducks
- The Language of Evolution
. Defining
Evolution
.
The
God of the Gaps
. Rube
Goldberg Machines
.
Evolving
the Irreducible
. Gregor
Mendel
.
Natural
Selection
. Computer
Evolution
.
The
Chicken or the Egg
. Antibiotic
Resistance
.
The
Immune System
. Pseudogenes
.
Genetic
Phylogeny
. Fossils
and DNA
.
DNA
Mutation Rates
. Donkeys,
Horses, Mules and Evolution
.
The
Fossil Record
. The
Geologic Column
.
Early Man
. The
Human Eye
.
Carbon
14 and Tree Ring Dating
. Radiometric
Dating
.
Amino Acid
Racemization Dating
. The
Steppingstone Problem
.
Quotes
from Scientists
. Ancient
Ice
.
Meaningful
Information
. The
Flagellum
.
Harlen Bretz
. Milankovitch
Cycles
Since
June 1, 2002




 Everyone
has experienced sickness at one time or another in his or her life.
However, most people get better and often do not get that same "bug" again.
Why is this? There are
literally millions of "bugs" and other things in our
everyday environment that can make us sick.
Why then do we generally remain so healthy?
The reason is because most of us have a highly effective immune system.
A healthy immune system is very good at searching out and killing foreign
invaders that can make the body sick. However, if the immune system is not
functioning well, there will certainly be a lot of problems.
Everyone
has experienced sickness at one time or another in his or her life.
However, most people get better and often do not get that same "bug" again.
Why is this? There are
literally millions of "bugs" and other things in our
everyday environment that can make us sick.
Why then do we generally remain so healthy?
The reason is because most of us have a highly effective immune system.
A healthy immune system is very good at searching out and killing foreign
invaders that can make the body sick. However, if the immune system is not
functioning well, there will certainly be a lot of problems.  Well,
T-cells are capable of being able to tell the difference between certain molecules (antigens) that are
presented (by MHC class I molecules) on
the surfaces of all "self" cells in the body that they are supposed to
protect compared to all foreign or "abnormal" antigens that are
associated with outside invaders or internal disease processes (like cancer).
Well,
T-cells are capable of being able to tell the difference between certain molecules (antigens) that are
presented (by MHC class I molecules) on
the surfaces of all "self" cells in the body that they are supposed to
protect compared to all foreign or "abnormal" antigens that are
associated with outside invaders or internal disease processes (like cancer). Not all the T-cells have different Y-shaped receptors,
but many of them do.
Not all the T-cells have different Y-shaped receptors,
but many of them do. The
B-cell that recognized the foreign antigen clones itself to make many
nearly identical copies of itself - with slight variations.
The
B-cell that recognized the foreign antigen clones itself to make many
nearly identical copies of itself - with slight variations.