.

|
Table of Contents |
The Debate
Dr. Jason
Rosenhouse is an associate professor of mathematics at James Madison University
in Virginia. In his spare time, he enjoys engaging in the debate over Creation
vs. Evolution and has even written an interesting book about his experiences
visiting creation events around the United States entitled, "Among
the Creationists." I personally enjoyed reading
this book very much, and found it to be very similar to my own experience
discussing this topic with both creationists and evolutionists alike. Most on
both sides of this issue simply aren't very well informed about arguments from
the opposing side, or even from their own side of the debate. I think reading
about these experiences is helpful, for those on both sides of this issue, to at
least start to understand how much we seem to be talking past each other, and to
understand the mindset and motivations of those within opposing camps.
 I
was asked to read Rosenhouse's book by Harry Allen, a radio host for "Nonfiction",
the on-air magazine of the arts, ideas, and fact in New York (airs Fridays,
2-3pm, on WBAI-NY/99.5 FM (wbai.org),
flagship of the non-commercial Pacifica radio network). He wanted me to have an
on-air
debate with Dr. Rosenhouse regarding his book
and the Evolution/Creation debate at large and why I'm involved with this debate
and have written my own book, "Turtles
All the Way Down." So, on Friday, January 17th,
we did just that (see Link below).
I
was asked to read Rosenhouse's book by Harry Allen, a radio host for "Nonfiction",
the on-air magazine of the arts, ideas, and fact in New York (airs Fridays,
2-3pm, on WBAI-NY/99.5 FM (wbai.org),
flagship of the non-commercial Pacifica radio network). He wanted me to have an
on-air
debate with Dr. Rosenhouse regarding his book
and the Evolution/Creation debate at large and why I'm involved with this debate
and have written my own book, "Turtles
All the Way Down." So, on Friday, January 17th,
we did just that (see Link below).
Jason Rosenhouse vs. Sean Pitman

I personally
enjoyed the discussion very much, and I think that Dr. Rosenhouse did as well
(given his description of our debate on his own
Blog). As far as a
summary of our discussion, Mr. Allen started us off with asking us something
about ourselves, how we became interested in the Creation/Evolution debate, and
why we wrote our books. He then asked me what I thought of Dr. Rosenhouse's
book and commented that he also really enjoyed reading the book. I explained
that I really enjoyed reading "Among the Creationists", that I read the whole
book in one sitting, and agreed with most of what Dr. Rosenhouse had to say, at
least 85% of it. Of course, Mr. Allen then asked me what parts I didn't agree
with...
 When
I first started reading, "Among the Creationists", I thought the book was going
to be more about the problems with standard Creationist arguments, and the
rational scientific reasons why evolution had to be true in light of these
arguments. I was especially intrigued by the fact that Dr. Rosenhouse is a
mathematician and the cover to the book explained that creationists get into
real trouble whenever they start to get into a discussion with Dr. Rosenhouse
within his own field of expertise - i.e., mathematics/probability arguments for
intelligent design. However, as I read through the book, I was disappointed to
discover that Dr. Rosenhouse had not included a single mathematical/probability
argument in favor of the creative potential of the evolutionary mechanism of
random mutations and natural selection. In fact, as is almost always the case
for modern neo-Darwinists, he claimed, in his book and during our debate, that
the modern Theory of Evolution is not dependent upon mathematical arguments or
statistical analysis at all - at least with regard to the creative potential of random mutations and natural selection. In this, he seemed to argue that his own field of
expertise is effective irrelevant to the discussion - that, "It is the wrong
tool to use." Beyond this, he also explained that he wasn't a biologist or a
geneticist and that any discussion of biology would require bringing in someone
with more expertise and knowledge of the field of biology than he had.
When
I first started reading, "Among the Creationists", I thought the book was going
to be more about the problems with standard Creationist arguments, and the
rational scientific reasons why evolution had to be true in light of these
arguments. I was especially intrigued by the fact that Dr. Rosenhouse is a
mathematician and the cover to the book explained that creationists get into
real trouble whenever they start to get into a discussion with Dr. Rosenhouse
within his own field of expertise - i.e., mathematics/probability arguments for
intelligent design. However, as I read through the book, I was disappointed to
discover that Dr. Rosenhouse had not included a single mathematical/probability
argument in favor of the creative potential of the evolutionary mechanism of
random mutations and natural selection. In fact, as is almost always the case
for modern neo-Darwinists, he claimed, in his book and during our debate, that
the modern Theory of Evolution is not dependent upon mathematical arguments or
statistical analysis at all - at least with regard to the creative potential of random mutations and natural selection. In this, he seemed to argue that his own field of
expertise is effective irrelevant to the discussion - that, "It is the wrong
tool to use." Beyond this, he also explained that he wasn't a biologist or a
geneticist and that any discussion of biology would require bringing in someone
with more expertise and knowledge of the field of biology than he had.
At this point I began to wonder why we were having a debate at all if his own field of expertise was, according to him, effectively irrelevant to the conversation and that he was not prepared to present arguments from biology or genetics regarding the main topic at hand - i.e., the potential and/or limits of the evolutionary mechanism of random mutations combined with natural selection.
Sequence Space
 Perhaps this is the main reason why Dr. Rosenhouse started to get rather
irritated and flustered in the second half of our debate when I started asking
him to explain how the evolutionary mechanism could reasonably and predictably
do what he claims it did - i.e., actually come up with new functional systems
beyond very low levels of functional complexity. In order to do this, of
course, the evolutionary mechanism would have to get across an extremely vast
ocean of non-beneficial sequence options at these higher levels of functional
complexity in order to somehow find the very very rare beneficial islands or
"steppingstones" in an otherwise very empty and very large ocean (i.e., many
times larger than the size of our entire universe). How could these extremely
rare beneficial steppingstones actually be discovered in a timely manner? After
all, as I explained during our discussion, the ratio of beneficial vs.
non-beneficial sequences decreases, exponentially,
with each increase in the level of functional complexity of the protein-based
system in question - an observation that holds true for every
language/information system that is known to us (to include the English language
system, Russian, German, Italian, Morse Code, computer code, DNA codes, etc.).
Given this exponential decline in the ratio of beneficial vs. non-beneficial,
how then can random mutations continue to find, via a truly random search
algorithm, new qualitatively novel beneficial sequences at higher and higher
levels of functional complexity within what anyone would consider to be a
reasonable amount of time?
Perhaps this is the main reason why Dr. Rosenhouse started to get rather
irritated and flustered in the second half of our debate when I started asking
him to explain how the evolutionary mechanism could reasonably and predictably
do what he claims it did - i.e., actually come up with new functional systems
beyond very low levels of functional complexity. In order to do this, of
course, the evolutionary mechanism would have to get across an extremely vast
ocean of non-beneficial sequence options at these higher levels of functional
complexity in order to somehow find the very very rare beneficial islands or
"steppingstones" in an otherwise very empty and very large ocean (i.e., many
times larger than the size of our entire universe). How could these extremely
rare beneficial steppingstones actually be discovered in a timely manner? After
all, as I explained during our discussion, the ratio of beneficial vs.
non-beneficial sequences decreases, exponentially,
with each increase in the level of functional complexity of the protein-based
system in question - an observation that holds true for every
language/information system that is known to us (to include the English language
system, Russian, German, Italian, Morse Code, computer code, DNA codes, etc.).
Given this exponential decline in the ratio of beneficial vs. non-beneficial,
how then can random mutations continue to find, via a truly random search
algorithm, new qualitatively novel beneficial sequences at higher and higher
levels of functional complexity within what anyone would consider to be a
reasonable amount of time?
Closely-Spaced Steppingstones
In response to what I consider to be a fundamental challenge for evolutionary theory, Dr. Rosenhouse made a very interesting claim that I've heard only a couple of times before in my debates a few years ago on Talk.Origins.org. Dr. Rosenhouse explained that while I was correct that the beneficial steppingstones in sequence space are very very rare indeed, and that the ratio of potentially beneficial sequences vs. non-beneficial sequences does in fact decrease in an exponential manner with each increase in the level of functional complexity (i.e, the minimum structural threshold requirements, which includes the minimum size and specificity requirements of more and more complex systems), that this concept is entirely irrelevant to the potential for evolutionary progress. He argued that regardless of what the beneficial ratio might or might not be at a particular level of functional complexity evolution could still make real progress in a reasonable amount of time (i.e., this side of trillions upon trillions of years). When I asked him to explain how this might be so, this is what he said in a nutshell:
 "Because
the very rare beneficial steppingstones are not randomly scattered around Lake
Superior, but are lined up in a straight line one right after the other, it is
easy to cross the lake from one shoreline to the other along a path of closely
spaced steppingstones..."
"Because
the very rare beneficial steppingstones are not randomly scattered around Lake
Superior, but are lined up in a straight line one right after the other, it is
easy to cross the lake from one shoreline to the other along a path of closely
spaced steppingstones..."
It is in this way, Rosenhouse argued, that one can walk from one side of a massive lake or ocean, regardless of its size or the overall rarity of steppingstones, all the way to the other side without having to get into the water and blindly swim around randomly in an effort to try to find another steppingstone. Therefore, the most important concept to understand is not the rarity of the steppingstones, but "how they are arranged in sequence space". So, it's the arrangement of the beneficial steppingstones, not their rarity, that is key to understanding the creative potential of the evolutionary mechanism. That is why such statistical arguments having nothing to do with understanding the potential and/or limits of the evolutionary mechanism - according to Rosenhouse.
This is a great argument! Why didn't I think of it before? After all, this proposal does actually solve, quite nicely, the statistical problems for the evolutionary mechanism. In fact, it's such a neat solution to the problem that it seems downright obvious - especially from the perspective of a mathematician who has apparently had little exposure to the real world of biology or genetics. Given some understanding of the real world of proteins and how the rare beneficial or even stable sequences are actually distributed in sequence space, one starts to see the real problem for Rosenhouse's hypothesis - i.e., it just doesn't match the real world. Unfortunately, science isn't just based on hypotheses and theories that work well on paper. It is based on theories that actually represent empirical reality in a testable potentially falsifiable manner. Now, it isn't that Rosenhouse's hypothesis isn't testable. It is testable. The problem is that it fails the test. It simply doesn't represent reality. In the real world that exists outside of Rosenhouse's very neat imagination, the distribution of potentially beneficial, or even stable, protein sequences with sequences space is known by experimental observations. And, contrary to Dr. Rosenhouse's wonderful solution to the problem, the distribution of real beneficial protein sequences is not a linear distribution, but a random, effectively uniform, distribution that becomes more and more so with each step up the ladder of functional complexity. At higher and higher levels of sequence space, as the total number of protein sequences within the space grows exponentially, the ratio of potentially beneficial vs. non-beneficial sequences decreases exponentially - and the distribution of these more and more rare beneficial steppingstones becomes more and more uniformly random in appearance. It's much like stretching a sticky sheet until it starts to get holes in it. As the sheet stretches, the holes get exponentially bigger relative to the remaining material of the sheet. Pretty soon, the bridges between areas of fabric start to break down and fall apart, leaving truly isolated islands of fabric (or "steppingstones") that are not connected to any other island (for a more detailed discussion of this particular problem see: Link).
That, in a nutshell, is the fundamental problem for the theory of evolution - it's mechanism simply isn't capable of going beyond very very low levels of functional complexity this side of a practical eternity of time (trillions upon trillions of years). And, the reason for this is the exponential decline in potentially beneficial options that random mutations are capable of evolving in a given span of time, combined with the non-linear or non-clustered distribution of these potentially beneficial sequences within sequence space.
This is a fundamental problem for evolution that natural selection simply can't solve. The reason for this is that, as explained during the debate, natural selection cannot look into the future. It can only select, in a positive manner, what is working to some beneficial advantage right now. Intelligence, on the other hand, can look into the future to see potential advantages to various combinations of sequences that natural selection could never envision - as Dr. Stephen Meyer explains:
 “[Intelligent]
agents can arrange matter with distant goals in mind. In their use of language,
they routinely ‘find’ highly isolated and improbable functional sequences amid
vast spaces of combinatorial possibilities.”
“[Intelligent]
agents can arrange matter with distant goals in mind. In their use of language,
they routinely ‘find’ highly isolated and improbable functional sequences amid
vast spaces of combinatorial possibilities.”
Circumstantial Evidence
What's left at this point is very frustrating for evolutionists because, without a viable mechanism, pretty much all there is to prop up neo-Darwinism are what Rosenhouse referred to in our discussion as "circumstantial evidence" (35:34 in the discussion), to include the very popular argument that complex biomachines (like the human eye or the bacterial flagellar motor) are poorly designed and are full of "flaws" - that no one, certainly no omnipotent God, would have designed these machines with so many obvious flaws. However, as far as I see things, the main problem with the "design flaw" arguments is that these arguments don't really have anything to do with explaining how the evolutionary mechanism could have done the job. Beyond this, they don't really rule out intelligent design either because, even if someone could make it better, that doesn't mean that an inferior design was therefore not the result of deliberate intelligence. Lots of "inferior" human designed systems are none-the-less intelligently designed. Also, since when has anyone made a better human eye than what already exists? It's like my four year old son trying to explain to the head engineer of the team designing the Space Shuttle that he and his team aren't doing it right. I dare say that until Dr. Rosenhouse or Richard Dawkins can produce something as good or better themselves, that it is the height of arrogance to claim that such marvelously and beautifully functional systems are actually based on "bad design".
Downhill Evolution
 Beyond
this, consider the fairly well-known arguments of Dr.
Kenneth Miller (cell
and molecular biologist at Brown University) who once claimed that the Type III
secretory system (TTSS: a toxin injector responsible for some pretty nasty
bacteria - like the ones responsible for the Bubonic Plague) is evidence against
intelligent design. How so? Miller argued that the TTSS demonstrates how more
complex systems, like the flagellar motility system, can be formed from more
simple systems, like the TTSS since its 10 protein parts are also contained
within the 50 or so protein parts of the flagellar motility system. The only
problem with this argument, of course, is that it was later demonstrated that
the TTSS toxin injector actually devolved from the fully formed flagellar
system, not the other way around (Link).
So, as it turns out, Miller's argument against intelligent design is actually an
example of a degenerative change over time - i.e., a form of devolution, not
evolution. Of course, devolution is right in line with the predictions of
intelligent design. Consider, for example, that it is fairly easy to take parts
away from a system while maintaining subsystem functionality. It is another
thing entirely to add parts to a system to achieve a qualitatively new higher
level system of function that requires numerous additional parts to be in a
specific arrangement relative to each other before the new function can be
realized to any selectable advantage. Such a scenario simply doesn't happen
beyond very low levels of functional complexity because the significant number
of non-selectable non-beneficial modifications to pre-existing systems within a
gene pool that would be required to achieve such a feat would take far far too
long - i.e., trillions upon trillions of years. (See the following video of a
lecture I gave on this topic - starting at 27:00):
Beyond
this, consider the fairly well-known arguments of Dr.
Kenneth Miller (cell
and molecular biologist at Brown University) who once claimed that the Type III
secretory system (TTSS: a toxin injector responsible for some pretty nasty
bacteria - like the ones responsible for the Bubonic Plague) is evidence against
intelligent design. How so? Miller argued that the TTSS demonstrates how more
complex systems, like the flagellar motility system, can be formed from more
simple systems, like the TTSS since its 10 protein parts are also contained
within the 50 or so protein parts of the flagellar motility system. The only
problem with this argument, of course, is that it was later demonstrated that
the TTSS toxin injector actually devolved from the fully formed flagellar
system, not the other way around (Link).
So, as it turns out, Miller's argument against intelligent design is actually an
example of a degenerative change over time - i.e., a form of devolution, not
evolution. Of course, devolution is right in line with the predictions of
intelligent design. Consider, for example, that it is fairly easy to take parts
away from a system while maintaining subsystem functionality. It is another
thing entirely to add parts to a system to achieve a qualitatively new higher
level system of function that requires numerous additional parts to be in a
specific arrangement relative to each other before the new function can be
realized to any selectable advantage. Such a scenario simply doesn't happen
beyond very low levels of functional complexity because the significant number
of non-selectable non-beneficial modifications to pre-existing systems within a
gene pool that would be required to achieve such a feat would take far far too
long - i.e., trillions upon trillions of years. (See the following video of a
lecture I gave on this topic - starting at 27:00):
SETI, Granite Cubes, and Useful Predictions
 Finally, of course, there is the standard argument that intelligent design isn't
science because it produces nothing useful - i.e., no useful predictions. Does
this mean that SETI science isn't a real scientific enterprise? What about
anthropology or forensic sciences, which are based on the scientific ability to
detect deliberate design behind various artifacts found in nature? How then is
it any different to apply the very same arguments used to detect design in these
various modern scientific disciplines in a universal manner? And, if artificial
features are also found within the DNA and/or protein-based systems of living
things, so be it! How long does one have to look for a mindless natural
mechanism to explain something like a highly symmetrical polished granite cube,
even if found on an alien planet like Mars, before it is recognized as a true
artifact of intelligent design? - regardless of if this conclusion is deemed to
be "useful" or not by this or that mathematician or scientist?
Finally, of course, there is the standard argument that intelligent design isn't
science because it produces nothing useful - i.e., no useful predictions. Does
this mean that SETI science isn't a real scientific enterprise? What about
anthropology or forensic sciences, which are based on the scientific ability to
detect deliberate design behind various artifacts found in nature? How then is
it any different to apply the very same arguments used to detect design in these
various modern scientific disciplines in a universal manner? And, if artificial
features are also found within the DNA and/or protein-based systems of living
things, so be it! How long does one have to look for a mindless natural
mechanism to explain something like a highly symmetrical polished granite cube,
even if found on an alien planet like Mars, before it is recognized as a true
artifact of intelligent design? - regardless of if this conclusion is deemed to
be "useful" or not by this or that mathematician or scientist?
 In this light, consider the fairly recent confirmation of a long-standing prediction of intelligent design regarding the likely key functionality of portions of non-coding or "junk-DNA". As it turns out, the more and more research that is done on non-coding DNA (DNA that does not code for proteins), the more and more functionality is being discovered. Such discoveries simply weren't predictable from the Darwinian perspective where many, such as Richard Dawkins in particular, had argued that non-coding DNA was evolutionary garbage or remnants of past trials and errors. In contrast, those favoring the intelligent design or even the creationist position had long argued that at least some proportion of non-coding DNA probably had beneficial functionality to one degree or another. And, it turns out that the predictions of intelligent design have proved true. It seems like protein-coding genes are like the bricks and mortar for a house while the blueprint for what type of house to build resides within the non-coding DNA.
In this light, consider the fairly recent confirmation of a long-standing prediction of intelligent design regarding the likely key functionality of portions of non-coding or "junk-DNA". As it turns out, the more and more research that is done on non-coding DNA (DNA that does not code for proteins), the more and more functionality is being discovered. Such discoveries simply weren't predictable from the Darwinian perspective where many, such as Richard Dawkins in particular, had argued that non-coding DNA was evolutionary garbage or remnants of past trials and errors. In contrast, those favoring the intelligent design or even the creationist position had long argued that at least some proportion of non-coding DNA probably had beneficial functionality to one degree or another. And, it turns out that the predictions of intelligent design have proved true. It seems like protein-coding genes are like the bricks and mortar for a house while the blueprint for what type of house to build resides within the non-coding DNA.
Along these lines, many creationists have highlighted the fairly recent published claims from the ENCODE human genome project. In 2012, the science journal Nature published a very interesting news item (ENCODE: The human encyclopaedia, Sept 5, 2012). This article reported on the ongoing human genome project called the "Encyclopedia of DNA Elements" or ENCODE project. The scientists at ENCODE made a very startling, and very controversial, claim - that at least 80% of our genome is functional to one degree or another!
Of course, many scientists have responded rather strongly against this 80% functionality number (Link). And, the truth of the matter is that, while non-coding DNA probably does represent the blue print for higher organisms, directing how protein-coding DNA functions to a significant degree, this does not necessarily or even likely mean that most non-coding DNA is actually required - or even useful. After all, some ferns and salamander have genomes the same size or smaller than the human genome, and other ferns and salamander have genomes 50 times the size of the human genome (the human genome is comprise of ~3.5 billion bases). For additional comparisons, consider that a chicken's genome contains about 1.3 billion bases, a clam about 3.2 billion, some frogs have 6.5 billion, and a lady bug genome has about 0.3 billion (~300 million) bases - similar to the genome of a Japanese pufferfish which is 8 times smaller than the human genome (just 385 million base pairs compared to 3 billion base pairs for humans), yet does just fine. It is clearly impossible to guess the genome size of an organism just by looking at its apparent "complexity" or "simplicity" or the number of protein-coding "genes" in the genome - which doesn't seem to correlate with the overall size of eukaryotic genomes. This curious fact is currently known as the C-value enigma.
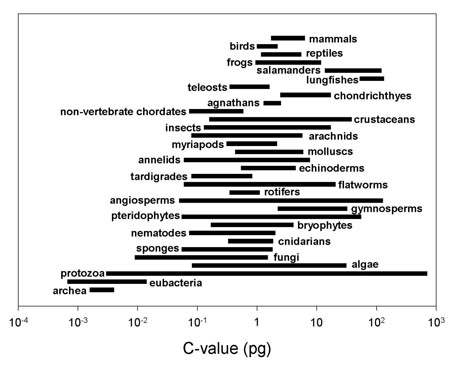
Given the reality of the C-value enigma, it seems likely that most non-coding DNA may not be vital for life or even beneficially functional. For example, consider the argument of Dr. Ryan Gregory known as "The Onion Test", with additional commentary from Dr. Larry Moran (Link):
The onion test is a simple reality check for anyone who thinks they have come up with a universal function for non-coding DNA. Whatever your proposed function, ask yourself this question: Can I explain why an onion needs about five times more non-coding DNA for this function than a human? The onion, Allium cepa, is a diploid (2n = 16) plant with a haploid genome size of about 17 pg. Human, Homo sapiens, is a diploid (2n = 46) animal with a haploid genome size of about 3.5 pg. This comparison is chosen more or less arbitrarily (there are far bigger genomes than onion, and far smaller ones than human), but it makes the problem of universal function for non-coding DNA clear. Further, if you think perhaps onions are somehow special, consider that members of the genus Allium range in genome size from 7 pg to 31.5 pg. So why can A. altyncolicum make do with one fifth as much regulation, structural maintenance, protection against mutagens, or [insert preferred universal function] as A. ursinum?
 However, there is the problem of the expense of maintaining stretches of DNA for long periods of time that don't provide any useful advantage to the organism. Such maintenance might seem to be fairly expensive if there is no return on the investment.
The counter argument, as presented by Dr. Nick Matzke, is that such maintenance really isn't very expensive at all relative to the other costs that the organism must pay. What's a few pennies here and there when you're spending thousands of dollars every day? Well, over millions of years a few pennies here and there might seem to add up to quite a lot. And, in a dog-eat-dog world, this could make all the difference. To add to the credibility of this observation, consider a paper by
Holloway
et al. (2007) where the authors observed a significant survivability cost disadvantage in various environments for bacteria that carried extra non-beneficial copies of DNA (Link).
However, there is the problem of the expense of maintaining stretches of DNA for long periods of time that don't provide any useful advantage to the organism. Such maintenance might seem to be fairly expensive if there is no return on the investment.
The counter argument, as presented by Dr. Nick Matzke, is that such maintenance really isn't very expensive at all relative to the other costs that the organism must pay. What's a few pennies here and there when you're spending thousands of dollars every day? Well, over millions of years a few pennies here and there might seem to add up to quite a lot. And, in a dog-eat-dog world, this could make all the difference. To add to the credibility of this observation, consider a paper by
Holloway
et al. (2007) where the authors observed a significant survivability cost disadvantage in various environments for bacteria that carried extra non-beneficial copies of DNA (Link).
However, the argument is that the cost is much higher for single-celled organisms compared to multi-celled organisms - like most eukaryotes. In this line, consider In 1980, two papers published in 1980 by Orgel and Crick (Link) and by Doolittle and Sapienza (Link) which that "selfish DNA" elements, such as transposons, essentially act as molecular parasites, replicating and increasing their numbers at the relatively slight expense of a host genome - so slight that natural selection simply can't keep up with the rate of expansion of these self-replicating elements within the genome.
However, a few scientists have suggested various functional options that might help to explain, to at least some degree, the C-value enigma. For example, non-coding DNA seems to act as a sort of clock to regulate the timing of expression of various genes and genetic elements during development (Swinburne, 2010). There is also the interesting discovery that the initiation of DNA replication and the transition from G1 to S is dependent upon nuclear volume. "Replication appears to initiate and terminate at the nuclear periphery and require a critical nuclear volume for onset (Nicolini et al., 1986); G1 nuclear volume growth must depend on concerted expansion of both chromatin and the nuclear envelope." (Cavalier-Smith, 2004) In short, "A genome's sheer bulk can influence the rate of cell division and thereby that of development." (Link)
Of course, the obvious counter is that such functionality for repetitive DNA is not dependent on the nature of the sequence itself, but only upon the absolute size of the sequence. And, while this appears to be true, having the right size in just the right place can obviously be quite beneficial. In other words, on occasion, size does matter...
This isn't all, of course. There are times when the actual specificity of the sequence matters as well - and this is what has also been discovered about non-coding DNA. It appears to be the blueprint that controls how the building blocks (i.e., the protein-coding genes) are used. In other words, non-coding DNA does seem to be more important than the protein-coding genes themselves. It seems, for instance, that it is the non-coding DNA that determines if a mouse or a pig or a monkey or a human is to be built given a set of very similar protein-coding genes for each of these types of creatures (for further discussion of this topic see: Link).
"I think this will come to be a classic story of orthodoxy derailing objective analysis of the facts, in this case for a quarter of a century," Mattick says. "The failure to recognize the full implications of this particularly the possibility that the intervening noncoding sequences may be transmitting parallel information in the form of RNA molecules - may well go down as one of the biggest mistakes in the history of molecular biology."
Wayt T. Gibbs, "The Unseen Genome: Gems Among the Junk," Scientific American (Nov. 2003).
. Home Page . Truth, the Scientific Method, and Evolution
.
. Maquiziliducks - The Language of Evolution . Defining Evolution
.
.
Evolving the Irreducible
.
.
.
.
.
. DNA Mutation Rates . Donkeys, Horses, Mules and Evolution
.
.
. Amino Acid Racemization Dating . The Steppingstone Problem
.
.
. Harlen Bretz . Milankovitch Cycles
. Kenneth Miller's Best Arguments
Since June 1, 2002