The Evolution of Mitochondria
Sean Pitman, MD
© February 2016
Biological Evolution:
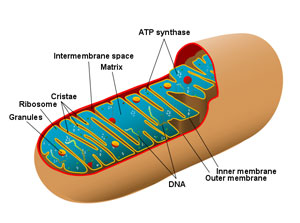
Many evolutionists often refer to an old argument that eukaryotic organisms (which includes humans) originally evolved hundreds of millions of years ago through a symbiotic relationship with bacteria. These internalized bacteria in turn eventually evolved into mitochondria – which are currently the source of the energy currency (in the form of the ATP molecule) of the eukaryotic cell.
Now, it is true that mitochondrial organelles are quite unique and very interesting. Unlike any other organelle, except for chloroplasts, mitochondria appear to originate only from other mitochondria. They contain some of their own DNA, which is usually, but not always, circular – like circular bacterial DNA (there are also many organisms that have linear mitochondrial chromosomes with eukaryotic-style telomeres). Mitochondria also have their own transcriptional and translational machinery to decode DNA and messenger RNA and produce proteins. Also, mitochondrial ribosomes and transfer RNA molecules are similar to those found in bacteria, as are some of the components of their membranes. In 1970, these and other similar observations led Dr. Lynn Margulis to propose an extracellular origin for mitochondria in her book, Origin of Eukaryotic Cells (Margulis, 1970). However, despite having their own DNA, mitochondria do not contain anywhere near the amount of DNA needed to code for all mitochondria-specific proteins. Over 99% of the proteins needed for mitochondrial function are actually produced outside of the mitochondria themselves. The DNA needed to code for these proteins is located within the cell’s nucleus and the protein sequences are assembled in the cytoplasm of the cell before being imported into the mitochondria (Endo and Yamano, 2010). It is hypothesized that these necessary genes were once part of the mitochondrial genome, but were then transferred and incorporated into the eukaryotic nuclear DNA over time. Not surprisingly then, none of the initial mtDNAs investigated by detailed sequencing, including animal mtDNAs, look anything like a typical bacterial genome in the way in which genes are organized and expressed (Michael Gray, 2012).
It is interesting to note at this point that Margulis herself wasn’t really very Darwinian in her thinking. She opposed competition-oriented views of evolution and stressed the importance of symbiotic or cooperative relationships between species. She also argued that standard neo-Darwinism, which insists on the slow accrual of mutations by gene-level natural selection, “is in a complete funk” (Link).
But what about all of those similarities between mitochondria and bacteria? It would seem like these similarities should overwhelmingly support the theory of common ancestry between bacteria and mitochondria.
 Well, the problem with Darwinian thinking in general is that too much emphasis is placed on the shared similarities between various creatures without sufficient consideration of the uniquely required functional differences. These required differences are what the Darwinian mechanism cannot reasonably explain beyond the lowest levels of functional complexity (or minimum structural threshold requirements). The fact of the matter is that no one has ever observed nor has anyone ever published a reasonable explanation for how random mutations combined with natural selection can produce any qualitatively novel protein-based biological system that requires more than a few hundred specifically arranged amino acid residues – this side of trillions upon trillions of years of time. Functionally complex systems that require a minimum of multiple proteins comprised of several thousand specifically-coded amino acid residue positions, like a rotary flagellar motility system or ATPsynthase (illustrated), simply don’t evolve. It just doesn’t happen nor is it remotely likely to happen in what anyone would call a reasonable amount of time (Link). And, when it comes to mitochondria, there are various uniquely functional features that are required for successful symbiosis – that bacteria simply do not have. In other words, getting a viable symbiotic relationship established to begin with isn’t so simple from a purely naturalistic perspective.
Well, the problem with Darwinian thinking in general is that too much emphasis is placed on the shared similarities between various creatures without sufficient consideration of the uniquely required functional differences. These required differences are what the Darwinian mechanism cannot reasonably explain beyond the lowest levels of functional complexity (or minimum structural threshold requirements). The fact of the matter is that no one has ever observed nor has anyone ever published a reasonable explanation for how random mutations combined with natural selection can produce any qualitatively novel protein-based biological system that requires more than a few hundred specifically arranged amino acid residues – this side of trillions upon trillions of years of time. Functionally complex systems that require a minimum of multiple proteins comprised of several thousand specifically-coded amino acid residue positions, like a rotary flagellar motility system or ATPsynthase (illustrated), simply don’t evolve. It just doesn’t happen nor is it remotely likely to happen in what anyone would call a reasonable amount of time (Link). And, when it comes to mitochondria, there are various uniquely functional features that are required for successful symbiosis – that bacteria simply do not have. In other words, getting a viable symbiotic relationship established to begin with isn’t so simple from a purely naturalistic perspective.
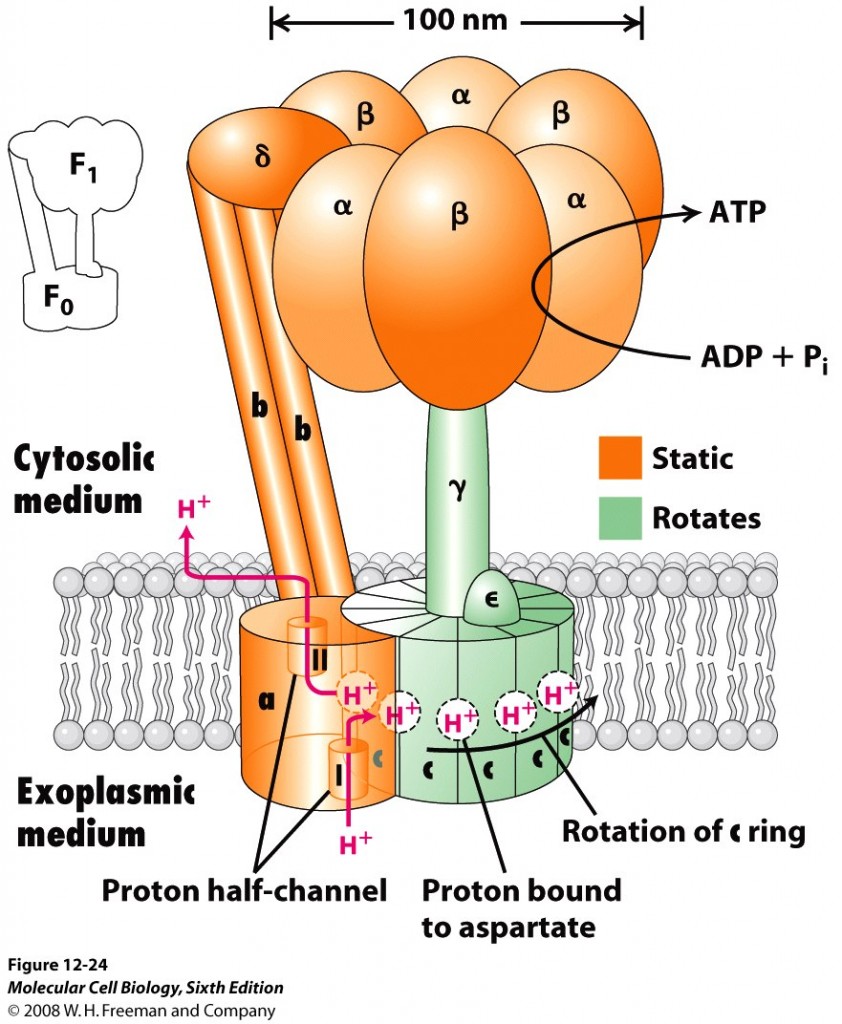 Consider, for example, that bacteria are not in the habit of sharing the ATP molecules they produce. Their ATPsynthase machine is set up so that the precious ATP molecules that are produced are released on the inside of the bacterium and the bacterium does not export the ATP that it makes. Of course, this same setup is also found in the mitochondria, but with one key difference. Mitochondria do export ATP for use by the host cell – but how? Now, both ATP and ADP (from which ATP is made) are highly charged molecules that do not diffuse freely across the inner mitochondrial membrane. So, how do the ATP molecules get outside of the mitochonria once they are produced so that they can be used by the host cell? Well, this requires a very specialized protein translocator protein (ATP/ADP Translocase; also known as Adenine nucleotide translocator or ANT) that moves the ATP molecules from the inner “matrix” of the mitochondria outward to the “intermembrane space” in exchange for ADP molecules (which are moved in the opposite direction). This system is set up so that the entry of ADP into the matrix is precisely coupled to the exit of ATP. ATP/ADP exchange is energetically expensive; about a quarter of the energy yield from electron transfer by the respiratory chain is consumed to regenerate the membrane potential that is consumed during this exchange process (Berg et al., 2002). And, this ANT machine is comprised of a homodimer with each subunit consisting of 297 residues and weighing approximately 30 kDa. Now that ATP is in the intermembrane space, it has to pass through another hurdle or gate-keeper channel known as the “voltage-gated channel” or VDAC (mitochondrial porin) – which is also a fairly complex in both structure and function (Dyson and Forte, 2008). As it turns out, ATP translocation through VDAC is facilitated by a set of specific interactions between ATP and the channel wall (Komarov, 2005).
Consider, for example, that bacteria are not in the habit of sharing the ATP molecules they produce. Their ATPsynthase machine is set up so that the precious ATP molecules that are produced are released on the inside of the bacterium and the bacterium does not export the ATP that it makes. Of course, this same setup is also found in the mitochondria, but with one key difference. Mitochondria do export ATP for use by the host cell – but how? Now, both ATP and ADP (from which ATP is made) are highly charged molecules that do not diffuse freely across the inner mitochondrial membrane. So, how do the ATP molecules get outside of the mitochonria once they are produced so that they can be used by the host cell? Well, this requires a very specialized protein translocator protein (ATP/ADP Translocase; also known as Adenine nucleotide translocator or ANT) that moves the ATP molecules from the inner “matrix” of the mitochondria outward to the “intermembrane space” in exchange for ADP molecules (which are moved in the opposite direction). This system is set up so that the entry of ADP into the matrix is precisely coupled to the exit of ATP. ATP/ADP exchange is energetically expensive; about a quarter of the energy yield from electron transfer by the respiratory chain is consumed to regenerate the membrane potential that is consumed during this exchange process (Berg et al., 2002). And, this ANT machine is comprised of a homodimer with each subunit consisting of 297 residues and weighing approximately 30 kDa. Now that ATP is in the intermembrane space, it has to pass through another hurdle or gate-keeper channel known as the “voltage-gated channel” or VDAC (mitochondrial porin) – which is also a fairly complex in both structure and function (Dyson and Forte, 2008). As it turns out, ATP translocation through VDAC is facilitated by a set of specific interactions between ATP and the channel wall (Komarov, 2005).
The problem, of course, is that it is hard to imagine much of an endosymbiotic advantage for a primitive eukaryotic cell engulfing a bacterium and keeping it around for a while if the bacterium wasn’t providing an immediate advantage. After all, if the bacterium isn’t sharing its own ATP, and it is using the host’s resources to generate that ATP, the relationship between them would seem to be more parasitic than symbiotic. And, as it turns out, the complex and highly specific ANT translocation machine is not found in any bacterial genome with the exception of parasitic Rickettsia and Chlamydia bacteria (Stephan Schmitz-Esser, 2004).
So, the big question is, where did the Rickettsia and Chlamydia bacteria get these genes for the ANT translocation system? – since no other bacteria have this system and these particular bacteria have lost most of the genes that other free-living bacteria have? Well, as it turns out, they got the specialized ANT genes by horizontal transfer from plants. In other words, they are “plastid homologues” unrelated to mitochondrial ANT genes (Kurland and Andersson, 2000). In fact, there are several other cases of acquisition of eukaryotic genes by bacteria – to include isoleucyl-tRNA synthetases whose acquisition from eukaryotes by several bacteria is linked to antibiotic resistance, and proteases that may be implicated in the pathogenesis of Chlamydia. (Koonin et al. 2001). In fact, the acquisition of eukaryotic genes by bacteria is a well-known phenomenon – especially parasites (Link).
So, how do the ANT-like genes (TLC genes) turn the Rickettsia and Chlamydia bacteria into parasites? – when mitochondria are not parasitic? Well, the Rickettsia and Chlamydiabacteria turn the process on its head so that instead of using the system to export ATP, they use it to import ATP from the host in exchange for exporting ADP – thereby sapping the host cell of energy.
In short, the Rickettsial-type system (from which mitochondria supposedly evolved) originally gained the necessary genes, not from previous evolution or from some other type of bacterial species, but from eukaryotic plants! In other words, eukaryotic organisms already had the necessary genes to begin with, which were then stolen by bacteria, allowing them to become parasitic. And, VDAC-type porins are also found in bacteria (Godbole, 2011), but, like ANT genes in bacteria, are often used for pathogenic purposes (Anne Müller, 2002).
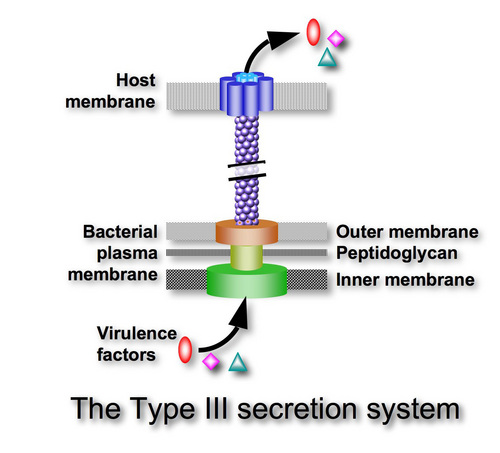
This is a form of degenerative evolution. It’s not a form of enhanced symbiotic functional complexity at all. In fact, there are many other examples of systems within parasitic organisms that originally arose, not by the evolution of simpler systems, but by the devolution or loss of informational complexity of more complex systems. Consider the Type III Secretory System (TTSS) used by pathologic bacteria as a toxin injector (bacteria such as Yersinia pestis, which are responsible for bubonic plague). For many years it was argued by prominent evolutionists (like Kenneth Miller or Nick Matzke) that the very functionally complex rotary bacterial flagellar motility system evolved from the TTSS system or something like it. It is now known, however, that the TTSS system devolved from the fully formed and functional flagellar motility system – not the other way around (Toft and Fares, 2008).
 Consider this in the light of the fact that almost all of the proteins used to build the rotary bacterial flagellum are homologous to one degree or another to other bacterial proteins that are doing other jobs as parts of other systems of function. However, homology alone doesn’t explain how all the modifications that are necessary to get all of these various proteins together to produce an entirely new and unique collective function. Until all of these required modifications are achieved, the next proposed beneficial steppingstone in the evolutionary pathway simply cannot be realized to any functional advantage that nature can select for in a positive manner (Link).
Consider this in the light of the fact that almost all of the proteins used to build the rotary bacterial flagellum are homologous to one degree or another to other bacterial proteins that are doing other jobs as parts of other systems of function. However, homology alone doesn’t explain how all the modifications that are necessary to get all of these various proteins together to produce an entirely new and unique collective function. Until all of these required modifications are achieved, the next proposed beneficial steppingstone in the evolutionary pathway simply cannot be realized to any functional advantage that nature can select for in a positive manner (Link).
The same problem is true for the mitochondrial system. Homologies don’t explain the necessary functional differences. Consider, for example, that around 42% of the proteins participating in the aerobic ATP-generating system of yeast mitochondria have been found only in eukaryotes (Kurland and Andersson, 2000). Also, “the protein translocases that drive protein import into mitochondria have no obvious counterparts in bacteria” (Dolzal, et al., 2006). And, these numerous required functional differences at these higher levels of functional complexity (I’ve only highlighted a couple of them here) are what help one detect the required signature of a very intelligent Designer behind these functionally complex systems.
How so? Because, beyond the lowest levels of functional complexity, biological systems, like any other functionally complex machine or functional computer code (where multiple specific orientations of building blocks or code are required) simply don’t evolve and are very unlikely to have ever evolved on this planet or anywhere else in the universe since the universe began. There’s just not remotely enough time, by anyone’s book, to make the odds favor the Darwinian mechanism when it comes to producing these higher level systems this side of a practical eternity of time. What’s left is just a story about how the evolution of this or that relationship must have happened - somehow. However, there’s really no science behind these fantastic stories - no viable mechanism much less any kind of meaningful demonstration, to back them up. It’s all based on the assumption that similarities between various living things must always mean common evolutionary ancestry – never common design.
. Home Page . Truth, the Scientific Method, and Evolution
.
. Maquiziliducks - The Language of Evolution . Defining Evolution
.
. Evolving the Irreducible .
.
.
.
.
. DNA Mutation Rates . Donkeys, Horses, Mules and Evolution
.
.
. Amino Acid Racemization Dating . The Steppingstone Problem
.
.
. Harlen Bretz . Milankovitch Cycles
. Kenneth Miller's Best Arguments